Our research projects
Exploring the virus diversity of vectors and wildlife at the crossroad of anthropogenic activity and climate change in Ecuador
Viral emerging and re-emerging diseases are a significant burden for global public health. These diseases are caused by either previously undetected or unknown viruses or known viruses that have spread to new geographic locations, new populations, or new host species. The geographic range of these pathogens is rapidly increasing and expected to further increase in the near future. Thereby, infectious viral diseases are spreading at a rate that has not been observed before. Due to climate change, globalization and land-use change, the probability increases that humans will come into close contact with wildlife species that are potential hosts or vectors of infectious virus increase. In addition, climatic conditions also have a significant impact on the local or regional emergence and frequency of zoonotic viruses, which is significantly influenced by the availability of potential host-species. However, the overall knowledge about the biodiversity of these viruses is very limited and almost unknown in South America. Joint integrated data analysis of host community compositions and virome composition will help to understand the spatiotemporal diversity and variability of virus transmission risk in different tropical and subtropical ecosystems and to estimate the potential changes in cause of the environmental shifts associated with climate change. The more we know about vector and wildlife viruses and the environmental drivers that may make them a potential threat for humans and livestock, the easier it will be for local communities and authorities to respond in a targeted informed with minimal impact to wildlife populations. This will allow disease-specific prevention and control measures to be applied in the case a specific virus becomes an emerging infectious disease.
The project aims to study the diversity of vectors (bats, rodents, mosquitoes) and associated viruses in the different tropical and subtropical areas of Ecuador, including remote tropical rainforest ecosystems populated by the indigenous (e.g. bush meat consumption). The areas are a biodiversity hotspot of the country, harboring a huge number of species in a small spatial area. Thus, the study on exploring vector and wildlife associated viruses along gradient of the different areas of Ecuador are a blueprint, which allows us to estimate the potential impact of climate change of an important host-virus interface worldwide. Therefore, will be an important contribution to assess the vulnerability and climate resilience of an important ecosystem in Ecuador.

Preparedness and Response in an Emergency context to Pathogens of MEDical and VETerinary importance
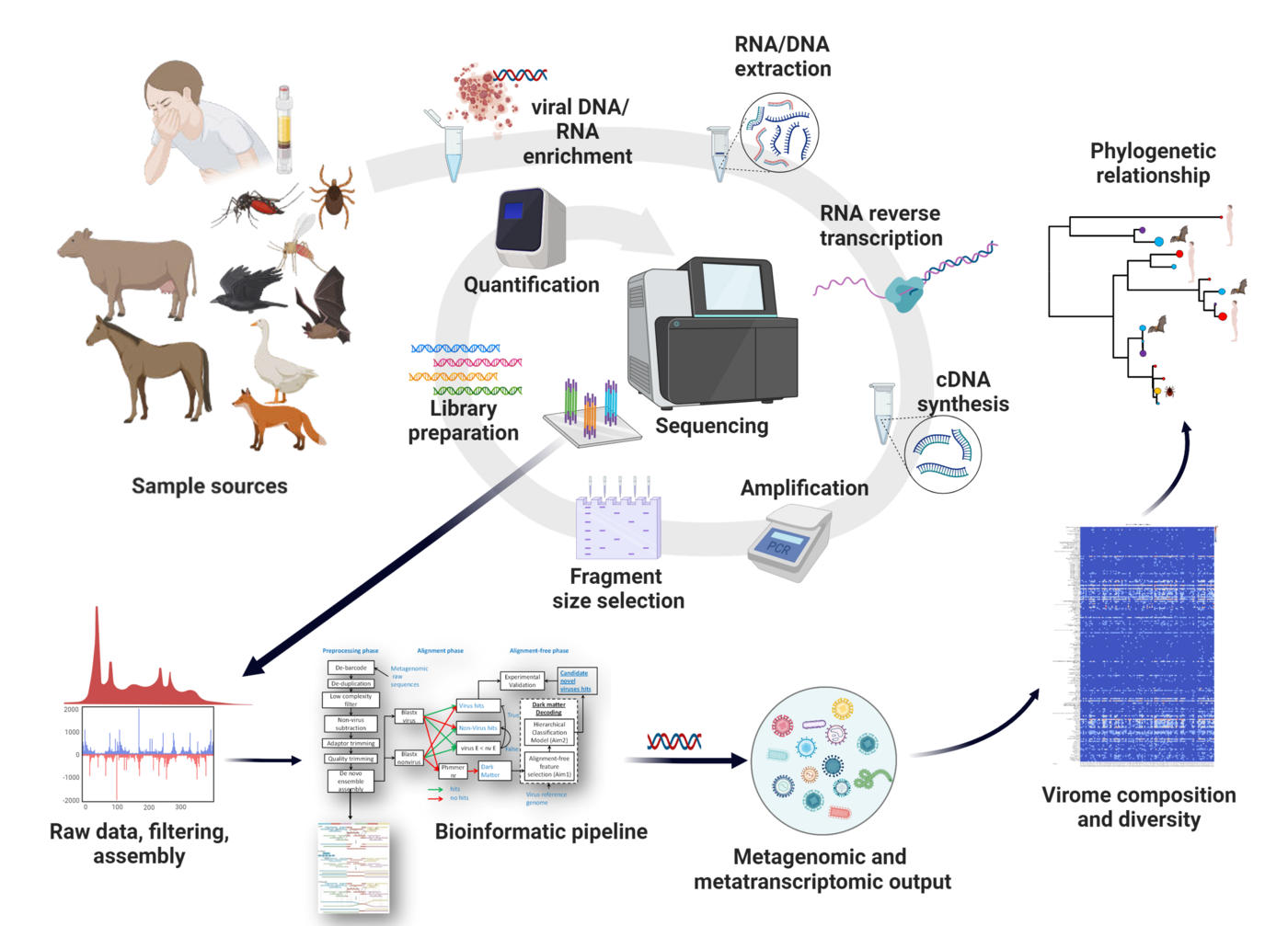
The rapid advancements in sequencing and computational technologies, coupled with a significant reduction in sequencing costs, have brought about a revolutionary shift in microbial genomics. These developments have provided unparalleled understanding of microbial evolution, diversity, and the interactions between hosts and pathogens. Additionally, genomic technologies have shown immense promise and suitability for clinical diagnostics and real-time detection and surveillance of epidemics.
The overarching objective of the PREPMedVet project is to establish a portable diagnostic sequencing platform (iSeq100, Illumina) capable of delivering critical information within a clinically relevant turnaround time during an outbreak. The consortium aims to develop a standardized wet-lab protocol that can be applied on-site using portable next-generation sequencing platforms. This approach will be complemented by user-friendly sequence analysis software, enabling the extraction of valuable insights into epidemic transmission patterns, pathogen genomics, evolution, and potential origins.
The project will focus on identifying pathogens and simultaneously produce and validate user-friendly detection assays. These assays can be stockpiled, transported, and distributed to end-users promptly, facilitating timely and comprehensive responses to outbreak scenarios. PREPMedVet is a BMBF (Federal Ministry of Education and Research) funded initiative.
High-throughput sequencing as a unique approach for xenosurveillance and high-resolution decoding of ecological vector-virus-host networks
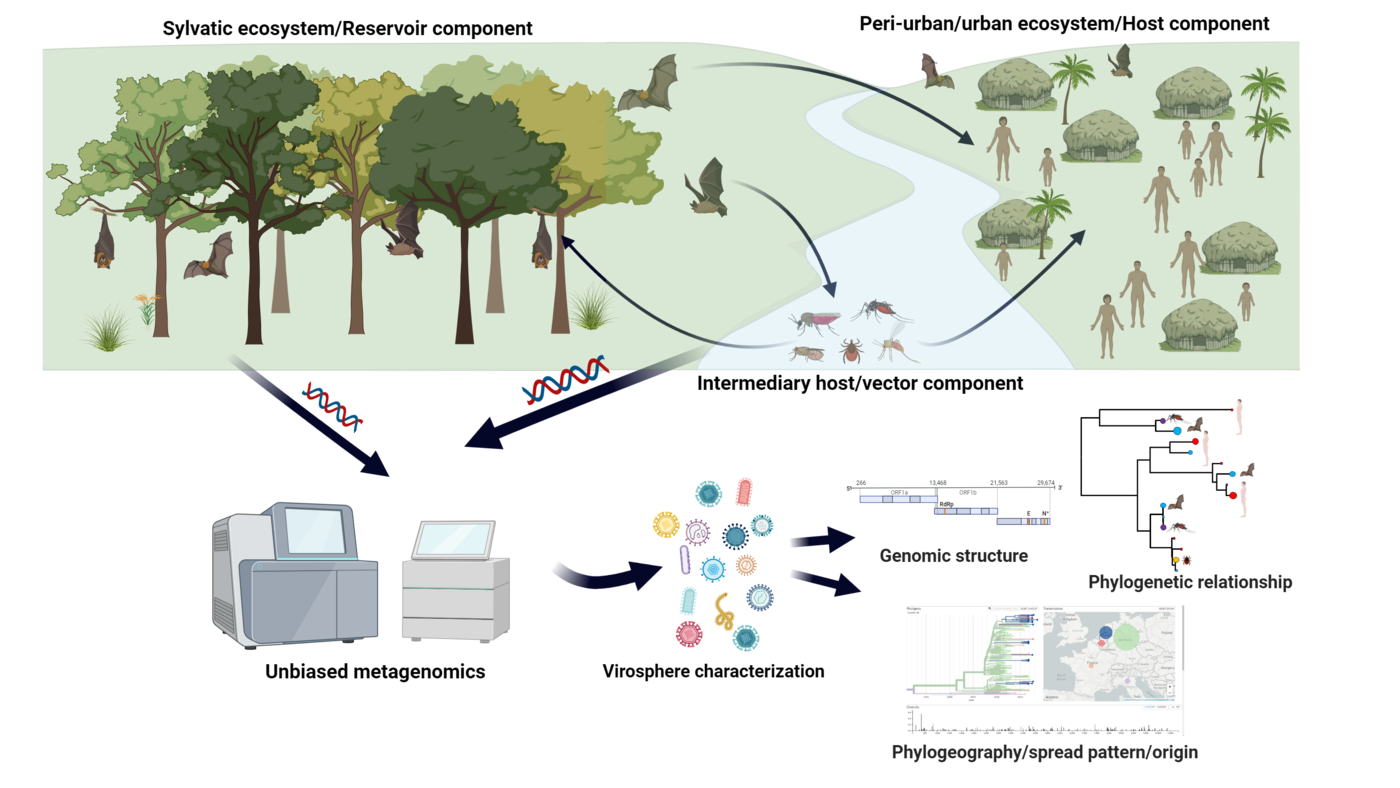
The spread of infectious agents by mosquitoes is a major cause of disease outbreaks affecting millions of people, livestock, and wildlife worldwide. There are several factors that have not been identified yet that could provide insights into the interplay between viruses, ecology, and mammalian health. Mechanisms that rely on that could increase awareness, identify the root causes of outbreaks, improve surveillance, and require rapid responses. There are different types of disease control that slow down or promote the spread of infectious diseases. "Xenosurveillance" is an approach to uncover the pathogenicity of diseases and their occurrence in an epidemiological context. Predicting vector-host interactions at the human-wildlife interface, such as the individual characteristics of these factors, are key elements for a better understanding of the uncontrolled events occurring. These predictions rely on high accuracy, which can be achieved using methods such as high-throughput sequencing. Broad surveys of viruses in nature are technically challenging, especially when aiming to cover taxa abundance and diversity at sufficient depth and within a spatiotemporal structure. An effective strategy is using massive parallel sequencing for the simultaneous identification of a wide range of viral types in subsamples of surveillance datasets. Since mosquito vectors feed on a wide range of animals and plants they effectively sample numerous viral reservoirs.
Understanding fundamental aspects of mosquito ecology is a prerequisite for discerning spatiotemporal patterns of arbovirus transmission and conceiving disease control strategies. In this context, we are interested in the characterization and diversity of their virome. Blood feeding is a defining step for the epidemiology of zoonotic pathogens. Mosquitoes feed on a large variety of vertebrate hosts, linking humans with other animals into vector-pathogen-host networks. Finding patterns of mosquito host selection is an essential aspect for understanding disease ecology and detection of unknown viruses with emergence potential. The use of genomic analysis has led to the identification of numerous biomarkers and to new insights into the interactions between the vector and the pathogen. However, so far there are no methods capable of characterizing the viral components, the host and the vector using next-generation sequencing. Therefore, the major objective is to develop a novel metagenomic/metatranscriptomoic method capable to decipher the virome composition of the vectors and decoding of ecological vector-virus-host networks. The Culifo3 is a BMEL (Federal Ministry of Food and Agriculture) funded project.

MOBILISE: A novel and green mobile One Health laboratory for (re-)emerging infectious disease outbreaks
MOBILISE envisages the creation of a cost-effective, novel BSL 3/4 mobile laboratory, which could be rapidly deployed in suspected outbreak sites, allowing a quick response at regional and national level. For most European countries this mobile laboratory will be the first to provide BSL3/4 capability and has the huge potential to dynamically increase overall testing capacity in the first phases of an outbreak. As most emerging viruses in Europe are zoonotic, the second important novelty of the MOBILISE laboratory is that it will be the first mobile laboratory to fully incorporate a One Health approach, by providing capacity to analyze human, animal, and environmental samples alike. This mobile laboratory prototype will provide up-to-date epidemiological data with minimal time-lag enabling accurate diagnosis, risk assessment and accelerated containment of disease by the responsible authorities.
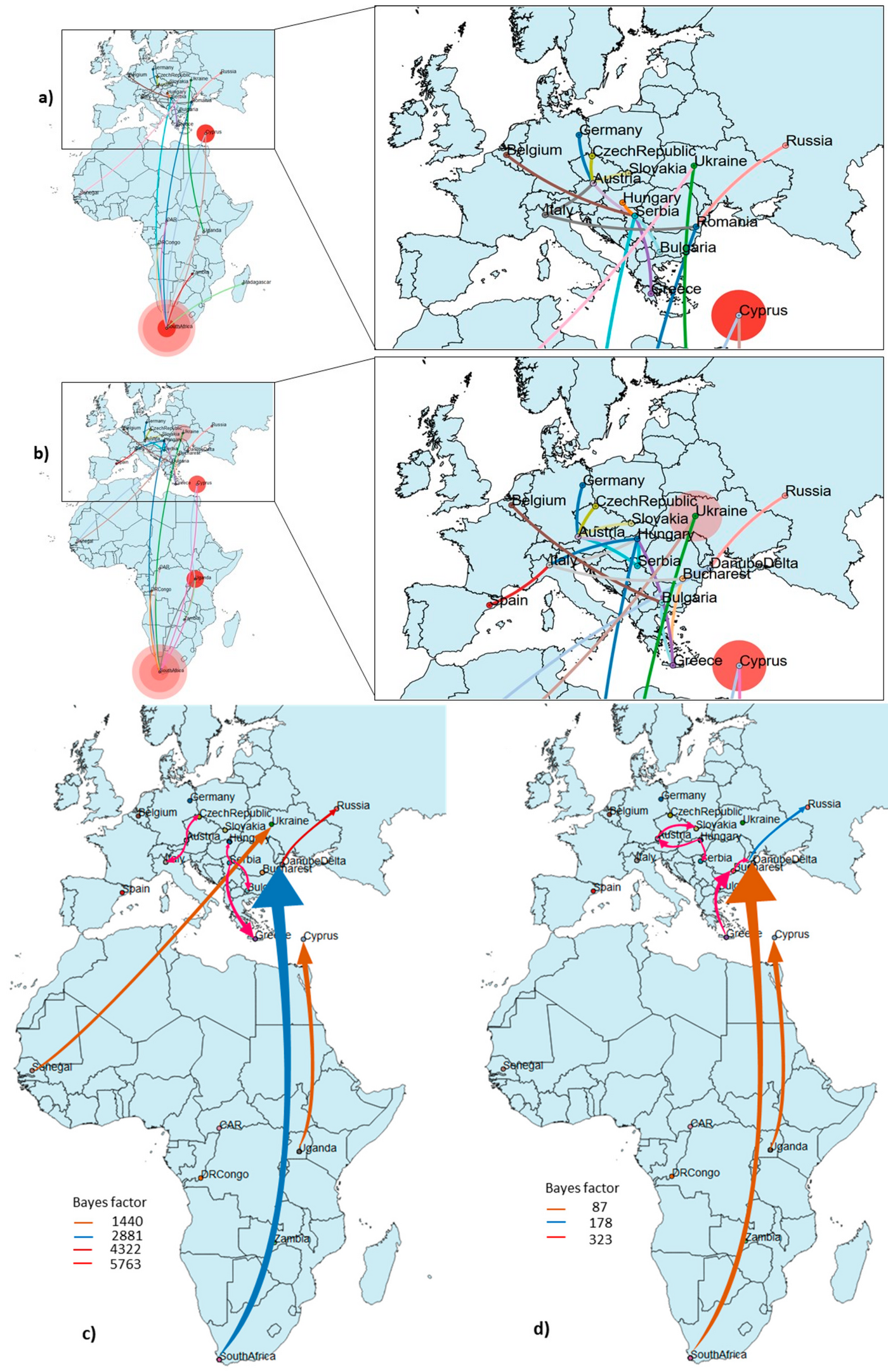
The main objective of our team within this Consortium is to implement a One Health Next-generation sequencing platform which will allow the identification of novel, highly divergent or unknown epidemic-prone viral pathogens (through an unbiased metagenomics screening approach) and the implementation of transmission networks in an outbreak. An established metagenomic and metatranscriptomic pipeline will be used and adapted to the smallest Illumina sequencing platform (iSeq100) to unravel the complete virome in the sample. The viral metagenomic and metatranscriptomic data obtained will be further used for 1) classification of the viruses by their phylogenetic patterns, 2) genomic and phylogenetic characterization of novel viruses, 3) classification of the viruses based on the host/vector species, 4) introduction and possible origin and 5) detection of potential hosts, prevalence, and epidemiology. The virome genetic variation and dynamics will be investigated by implementing Bayesian and maximum likelihood phylogenetic approaches. Layering additional climatic, transportation geographic, economic, and demographic information into a large phylogenetic data set will reveal the risk factors that facilitate local, regional or global spread. MOBILISE is a Horizon Europe (Europen Commission) funded project.

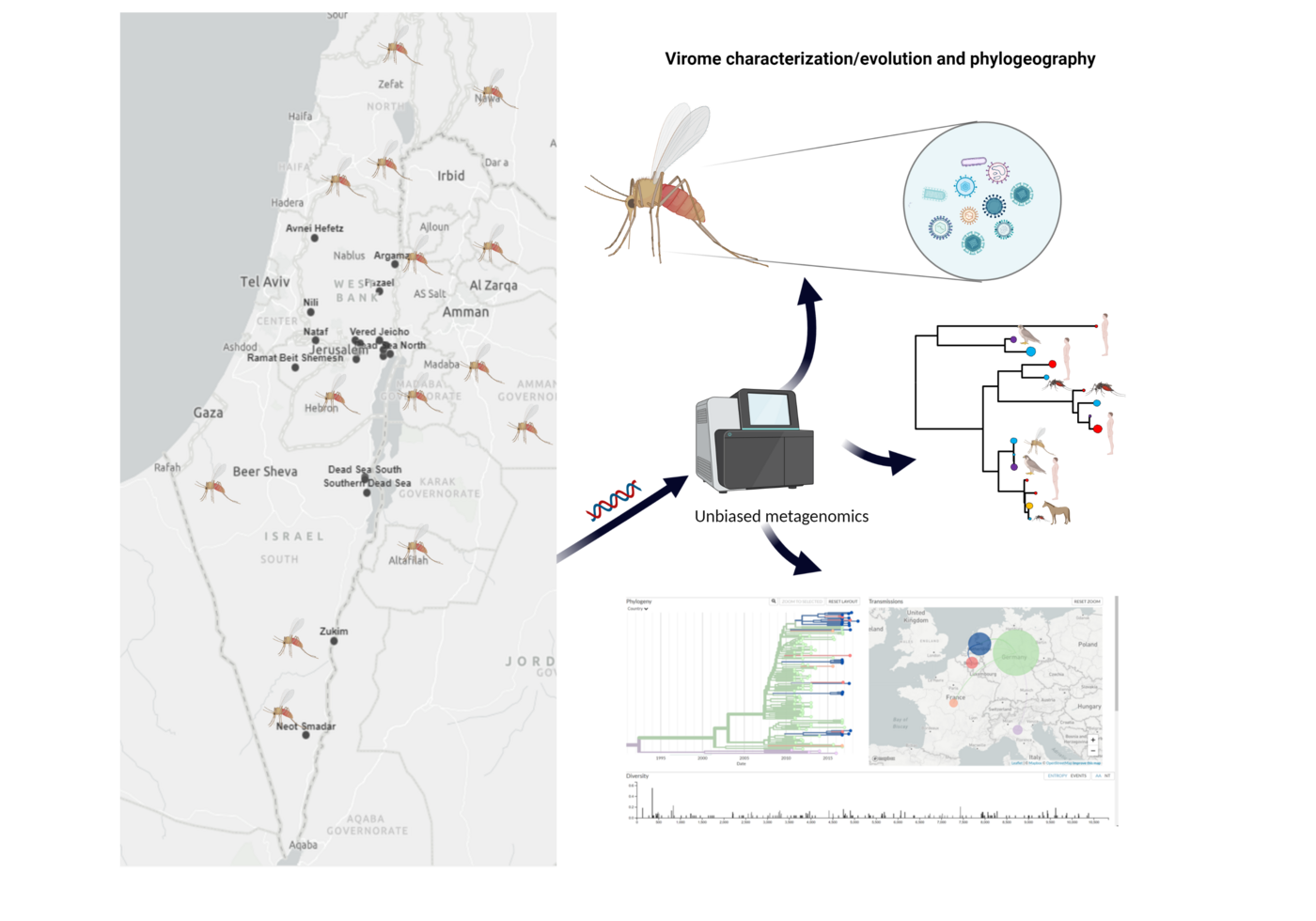
Sandfly-Borne Viruses from Israel and their Impact on Human and Animal Health
Mosquito-borne viruses, such as Zika, chikungunya and dengue caused several major epidemics and were extensively studied. In contrast, sand flies are also known to be vectors of relevant viral pathogens such as Toscana and Naples virus but the knowledge on evolution, ecology and medical importance are very scarce, especially in Israel. It is widely accepted that vector-borne viruses will continue to emerge and have an important impact on human health. Several sandfly-borne viruses mainly belonging to the Phlebovirus, Vesiculovirus and Orbivirus genera, are known to be pathogens for humans and animals in several countries worldwide.
Yet, identification of these viruses and assessment of their potential risk to public health is complex and requires the combination of viral metagenomics, entomological, environmental analysis, and clinical diagnosis. In this project we will use a novel approach to characterize the diversity of RNA/DNA viruses infecting sandflies known to transmit important zoonotic and animal viral diseases and to investigate the possible effects of this diversity on host-pathogen interactions in different areas of Israel. The study of viral diversity is important to understand the biology of any single viral species within an ecosystem and to help design strategies to control specific interactions between organisms. To date no sandfly borne virus from Israel has been isolated in cell culture or sequenced. The goal of the proposed research is to characterize known and new viruses in field collected sandflies of different species from diverse ecological environments and to determine whether the novel viruses represent potential risk for public health.
To achieve this objective, we will first set up country wide sandfly surveillance adjusted for virus detection and identify the relevant vector sandfly species, their host feeding patterns and spatial and temporal distribution in Israel. Next, we will examine the viral metagenomics found in sandflies collected in the field and characterize the viruses that might be associated with human disease. The identified novel and known sandfly-borne viruses will be isolated and serological tests will be developed. Finally, we will evaluate the functional and clinical relevance of the identified sand fly borne viruses by a seroprevalence cross sectional study of the human population in Israel. We expect to identify novel and known viruses that are transmitted by sandflies in Israel and elucidate the medical importance of such viruses in Israel.
Vector range, genetic traits and phylodynamic/phylogeography of arthropod-borne viruses in Danube Delta
The Danube Delta is the largest continuous marshland and the second largest and best-preserved river delta in Europe. The Danube Delta also represent the place with the highest biodiversity and biggest wetland in Europe and provides perfect environmental conditions necessary for a diverse and abundant mosquitoes fauna. Being located along a major migratory route, the Danube Delta represents an extremely important nesting and feeding zone for a large number of resident and migratory birds from Africa and Asia. Mosquito-borne diseases are characterized as being mostly local and focal with transmission foci and hotspots which are influenced by the presence of the aquatic biotopes suitable for the development of mosquito populations in the areas where blood feeding occurs. Wild birds are important to public health because they can act as biological or mechanical carriers of emerging zoonotic pathogens either as a reservoir host or by dispersing infected arthropod vectors. Bird migration provides the necessary factors for the establishment of new endemic foci of disease (maintained through a local bird–arthropod life cycle) even at great distances from where the pathogen was acquired. Given that the introduction and spread of the mosquito-borne viruses into a new geographic area depends on the presence of infected hosts, suitable mosquito vector and favorable habitats, the knowledge of the resident mosquito species population distribution and dynamics, their feeding preferences and vector competence are crucial to understand the epidemiological cycle and is useful in helping to predict future outbreaks of infections due to emerging zoonotic pathogens. In addition, a longitudinal mosquito surveillance will help us to learn more about mosquito-borne pathogens ecology and their genetic and phylogeographic characterization.