Understanding the pathogenicity of Entamoeba histolytica
The pathogen Entamoeba histolytica can live asymptomatically in the human gut, or it can disrupt the intestinal barrier and induce life-threatening abscesses in different organs, most often in the liver. The molecular framework that enables this invasive, highly pathogenic phenotype is still not well understood. In order to identify factors that are positively or negatively correlated for invasion and destruction of the liver, we used a unique tool, E. histolytica clones that differ dramatically in their pathogenicity, while sharing almost identical genetic background. Based on comprehensive transcriptome studies of these clones, we identified a set of candidate genes that are potentially involved in pathogenicity.
Overexpression and silencing transfectants will be used to analyse the influence of putative pathogenicity factors on the virulence of E. histolytica.
Furthermore, the invasion process will be analysed in detail using clones of different pathogenicity and an intestinal organoid-derived monolayer model. In addition, the role of extracellular vesicles secreted by E. histolytica in communication with the immune system will be investigated.
Organoid-derived monolayer model to study host-parasite interactions
Juliett Anders
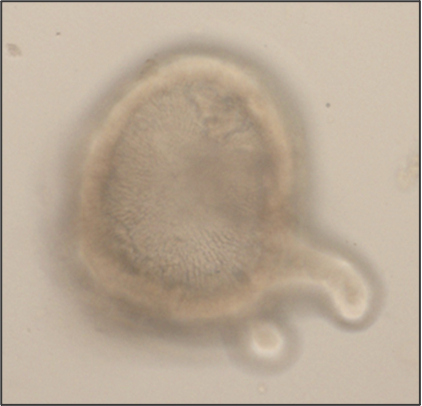
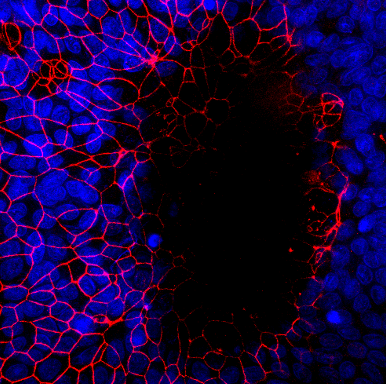
Intestinal organoids are in vitro systems, that mimic organ-specific structural and functional properties. These “mini-guts” possess a lumen and contain characteristic cell types of the intestinal tissue. To use these advantages in a model that is easily accessible for parasitic infections on the luminal side, an intestinal organoid-derived monolayer model was established in our lab.
The large intestinal epithelium is the location of invasion of the human pathogen Entamoeba histolytica. The monolayer model generated from large intestinal organoids is used to study the interaction of the parasite with the intestinal cells. In the future immune cells and a microbiome can be added to the system.
Funding: Deutsche Forschungsgemeinschaft (DFG).
Extracellular vesicles as communicators between parasite and host immune system
Barbara Honecker
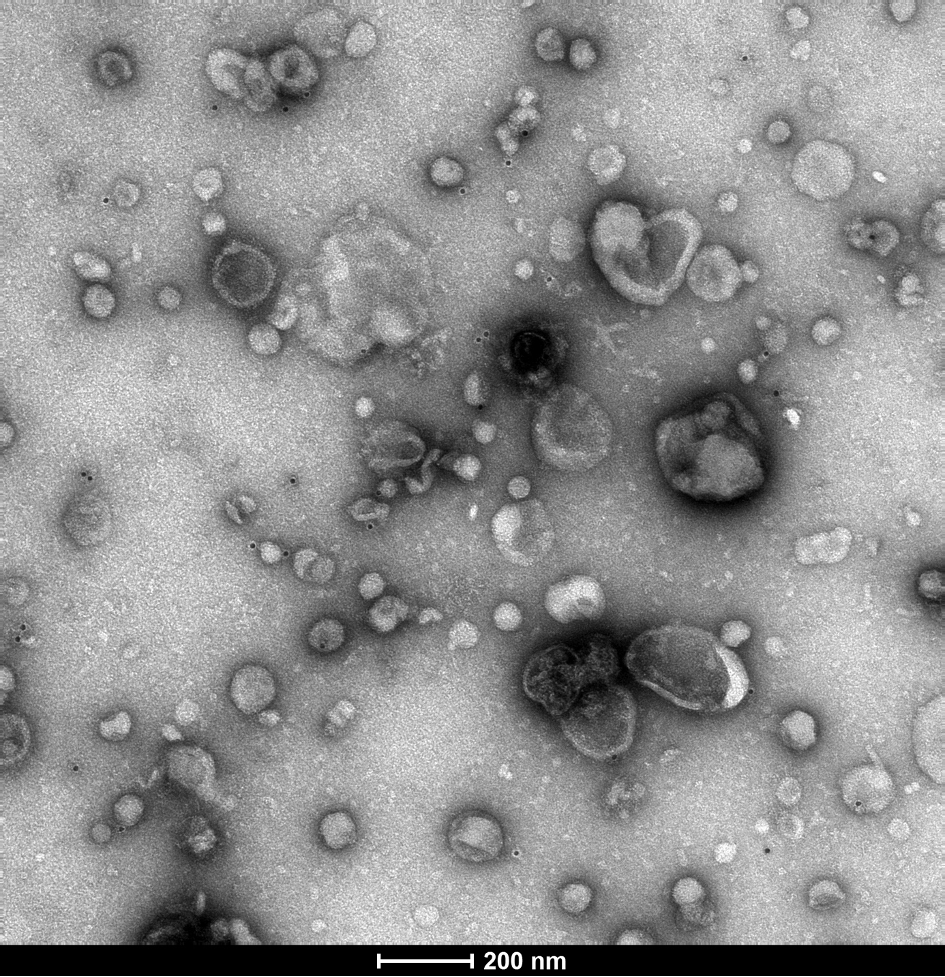
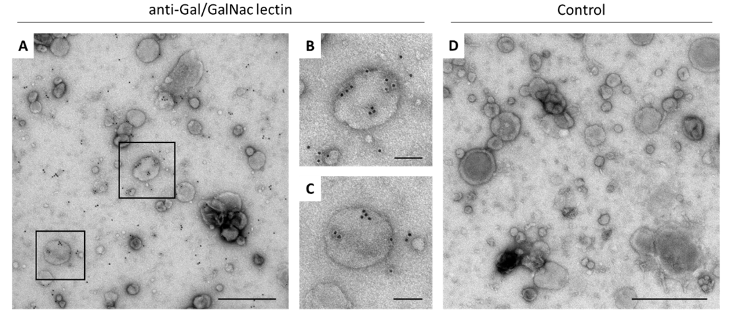
Extracellular vesicles (EVs) are small particles released by cells. For many parasitic disease models, it is known that EVs can play a role in the interaction between parasite and host and contribute to immune responses.
In this project, we investigate EVs secreted by Entamoeba histolytica, causative agent of the disease amebiasis. EVs are analyzed with regard to their biological properties as well as their immune-stimulatory capacities. The protein and miRNA cargo of EVs released by amoeba differing in their pathogenicity is characterized using mass spectrometry and miRNA sequencing. Differences between the protein and miRNA cargo of the two clones are investigated to gain further insight into pathogenicity mechanisms of the parasite.
In order to determine to which extent EVs play a role in communication with the host immune system, EVs isolated from parasite cultures are used to stimulate monocytes and neutrophils in vitro. Both immune cell types are known to be involved in the immune pathology of amebiasis. Immune cells derived from both male and female mice are used in order to investigate putative sex differences in response to EV stimulation.
Funding:
Graduate School‚ Infection Biology of Tropical Pathogens‘, Joachim Herz Foundation. Joint PhD project with research group Molecular Infection Immunology (AG Lotter).