Forschungsprojekte
Untersuchung der Replikationsmaschinerie von Bunyaviren
Die Ordnung der Bunyavirales ist eine äußerst vielfältige Gruppe von Viren, zu der zahlreiche humanpathogene Erreger gehören. Die WHO betont den dringenden Bedarf an medizinischen Gegenmaßnahmen gegen mehrere Bunyaviren, darunter das Lassa-Virus, das Virus des hämorrhagischen Krim-Kongo-Fiebers und das Rifttal-Fieber-Virus. Diese Viren stellen eine Bedrohung für die globale Gesundheit und die Wirtschaft dar, insbesondere in Ländern mit niedrigem und mittlerem Einkommen, aber sie sind auch für ihr hohes epidemisches Potenzial und, aufgrund des Klimawandels, ihre zunehmende geografische Verbreitung bekannt. Viele dieser Viren müssen in Laboren mit höchsten Biosicherheitsstandards gehandhabt werden, was das Methoden- und Technologiespektrum für die Forschung einschränkt. Trotz der schweren Krankheiten, die sie verursachen können, sind Bunyaviren faszinierend einfach: Sie enthalten ein segmentiertes Negativstrang-RNA-Genom (Erbinformation) und produzieren nur eine Handvoll eigener Proteine, manche kommen mit nur vier viralen Proteinen aus.
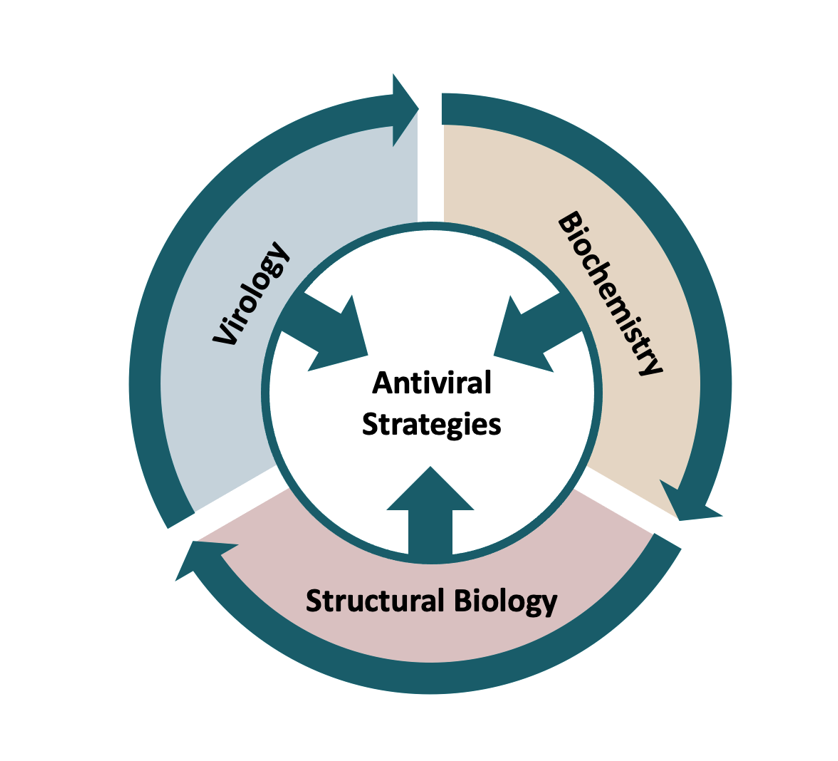
Zwei Proteine stehen hauptsächlich im Fokus unserer Forschung: Das große (L-) Protein und das Nukleoprotein N, welche zusammen mit dem RNA-Genom des Virus die Ribonukleopartikel bilden, die minimale Einheit für die Replikation und Transkription des viralen Genoms. Wir wenden eine Vielzahl von Methoden aus den Bereichen Strukturbiologie, Biochemie und Virologie an, um die Replikations- und Transkriptionsprozesse des Bunyavirus-Genoms in kleinsten Details zu verstehen. Diese Prozesse führen zur Produktion neuer viraler Proteine in den Zellen (virale Transkription) sowie zur Vervielfältigung des viralen Genoms (Genomreplikation), um schließlich neue virale Partikel zusammenzusetzen, die bereit sind, die nächsten Zellen zu infizieren. Der Hauptakteur in diesen Prozessen ist das virale L-Protein, eine multifunktionale und hoch optimierte molekulare Maschine, vergleichbar mit einem Schweizer Taschenmesser. Unser Ziel ist es, diese molekulare Maschine - ihre Funktionen, ihre Regulierung, ihre Interaktionen mit anderen Proteinen - zu verstehen, um schließlich Strategien zu entwickeln, wie die Prozesse der Genomreplikation und Transkription gehemmt und damit die Vermehrung des Bunyavirus gestoppt werden kann.
Im Folgenden stellen wir unsere Forschungsfragen und ausgewählte Veröffentlichungen etwas genauer vor.
Der Cap-Snatching-Mechanismus der Bunyaviren
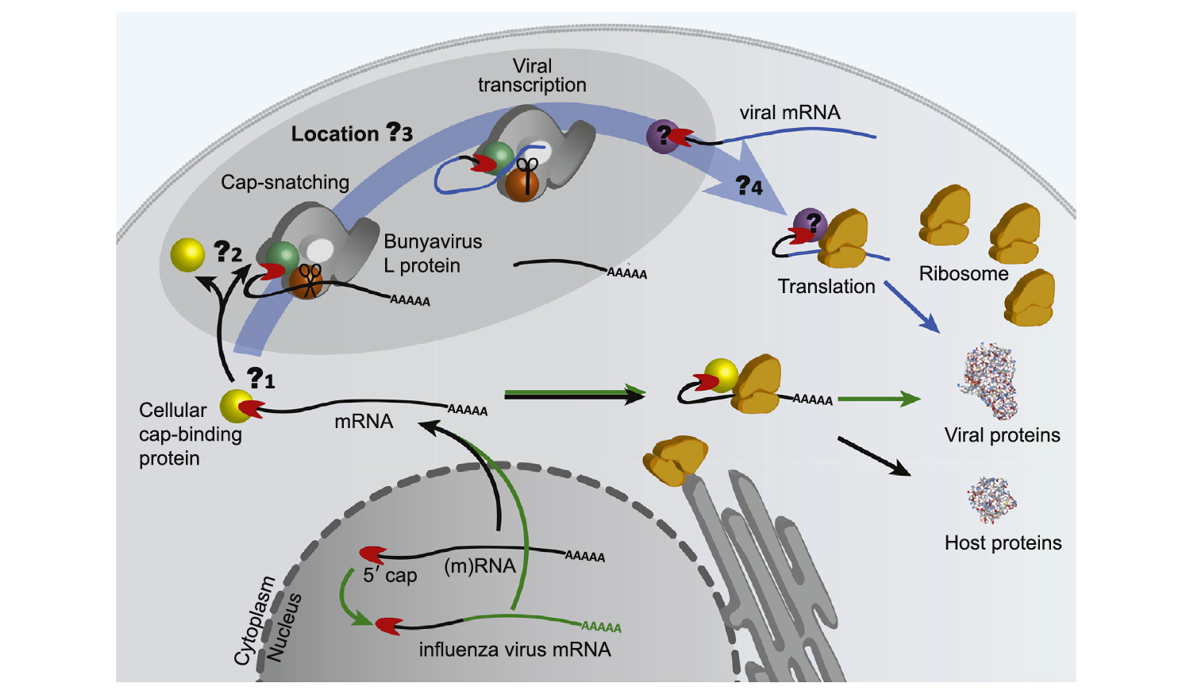
Viren haben verschiedene Strategien entwickelt, um ihre viralen Proteine in der Wirtszelle zu produzieren. Bunyaviren verwenden einen Cap-Snatching-Mechanismus, um die virale Transkription zu initiieren - das heißt sie stehlen sogenannte RNA-Kappen der Wirtszelle um die Übersetzung der viralen genetischen Information in virale Proteine durch die Wirtszelle zu erreichen. Beim Cap-Snatching-Mechanismus wird eine kurze Nukleotidsequenz inkl. RNA-Kappe vom 5'-Ende der Wirtszell-mRNA abgespalten und als Startpunkt der viralen mRNA-Produktion verwendet. Während dieser Prozess für Influenzaviren recht gut verstanden ist, ist unser Wissen über den genauen Mechanismus bei Bunyaviren noch sehr begrenzt.
In 2020 haben wir einen Übersichtsartikel veröffentlicht, welcher einen Überbick über die jüngsten strukturellen und funktionellen Daten zum Cap-Snatching-Mechanismus von Bunyaviren gibt. Wir haben Ähnlichkeiten und Unterschiede zwischen den im Nukleus replizierenden Influenzaviren und den im Zytoplasma replizierenden Bunyaviren beschrieben und noch offene Fragen zum Cap-Snatching-Mechanismus von Bunyaviren hervorgehoben: (1) Von welchen zellulären mRNAs wird das 5’-Cap gestohlen? (2) Wie binden Bunyaviren die RNA-Kappe? (3) Wo genau im Zytoplasma stehlen Bunyaviren die mRNA-Kappen? (4) Wie stellen Bunyaviren sicher, dass die viralen mRNAs an den Ribosomen der Wirtszelle in virale Proteine übersetzt werden?
Identifizierung einer funktionstüchtigen Cap-bindenden Domäne im Rifttal-Fieber-Virus L protein
Beim Prozess des Cap-Stehlens (Cap-snatching) wird ein kurzes, mit einem 5’-Cap versehenes Stück der Wirtszell-mRNA abgespalten und als Start der viralen mRNA-Synthese genutzt. Somit wird sowohl eine Endonuklease Domäne für die RNA-Spaltung benötigt als auch eine Cap-bindende Domäne, welche die Cap-Struktur der Wirtszell-mRNA bindet. Während der Cap-Snatching-Mechanismus für Influenzaviren bereits umfangreich untersucht wurde und beide Funktionen gefunden wurden, sind beim bunyaviralen Cap-Snatching-Mechanismus noch viele Fragen offen. Obwohl für die Bunyaviren eine Endonuklease bereits identifiziert wurde, konnte lange keine funtkionierende Cap-Bindungsaktivität festgestellt werden. In den vergangenen Jahren ist es uns gelungen durch Kristallstrukturen und biophysikalische Methoden die Existenz einer funktionstüchtigen Cap-Bindungsdomäne im bunyaviralen L-Protein zu beweisen. Die Cap-Bindungsdomäne der Bunyaviren ist dabei vom Aufbau her sehr ähnlich zur Influenzavirus Cap-bindenden Domäne mit einem zentralen β-Faltblatt über dem eine lange α-Helix verläuft. Die RNA-Kappe wird dabei zwischen zwei aromatischen Seitenketten gebunden.
Publikation:
Olschewski-Pavlita S, Cusack S, Rosenthal M. The Cap-Snatching Mechanism of Bunyaviruses. Trends Microbiol. 2020;28(4):293-303. doi:10.1016/j.tim.2019.12.006
Eine RNA sie zu knechten: Kritische Schritte bei der Bildung und Rekrutierung des Lassa-Virus Ribonukleoprotein-Komplexes
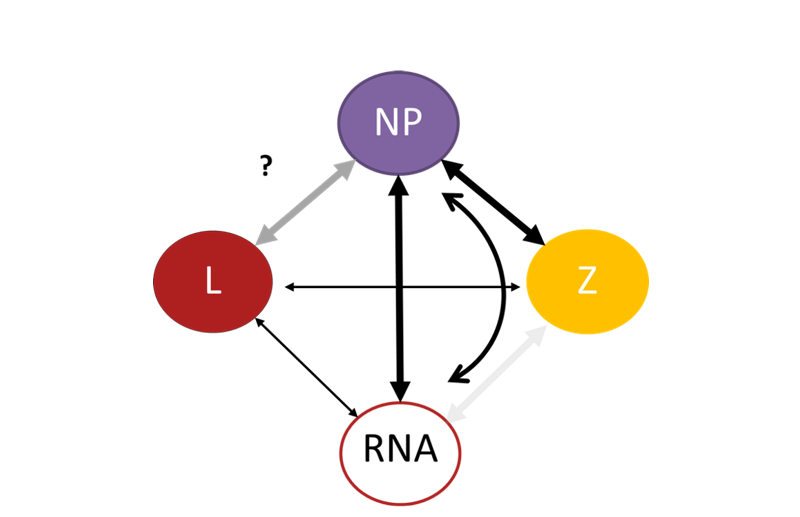
Virale Proteine müssen untereinander, mit Proteinen der Wirtszelle sowie mit viraler RNA und anderen Biomolekülen gezielt interagieren, um eine erfolgreiche Vermehrung des Virus zu gewährleisten. Wir verwenden die native Massenspektrometrie als Methode zur Analyse von Protein-Protein- und Protein-RNA-Wechselwirkungen in Bunyaviren. Diese Methode erlaubt es, Proteine in ihrem nativen Zustand zu "wiegen". Wir wollen verstehen, wie virale Ribonukleopartikel (RNP) zusammengebaut und für den Zusammenbau neuer Viruspartikel rekrutiert werden. Das RNP ist ein wichtiger Bestandteil des Virus, da es die minimale Einheit für die virale Genomreplikation und Transkription in Zellen darstellt (siehe oben). Wir haben entdeckt, dass das virale Nukleoprotein N, wenn es auf kleine RNA-Fragmente trifft, seine Form verändert und mit anderen N-Proteinen interagiert. Der ursprünglich trimere, ringförmige N-Komplex zerfällt in Monomere, die an die RNA gebunden sind. Diese an die RNA gebundenen N-Proteine bilden dann größere Komplexe, die an RNPs erinnern. Darüber hinaus haben wir festgestellt, dass ein anderes virales Protein, das so genannte Z-Protein, sowohl in Gegenwart als auch in Abwesenheit von RNA direkt mit N interagiert. Diese Wechselwirkung ist wichtig für die Rekrutierung von RNPs in neu entstehende Viruspartikel. Interessanterweise ist die Stärke dieser N-Z-Wechselwirkung in einem sauren Milieu geringer, ähnlich den Bedingungen, unter denen das Virus in unsere Zellen eindringt, was die Freisetzung der RNPs aus der Virushülle bei der Infektion neuer Zellen erleichtern könnte. Das Verständnis dieser Mechanismen der RNP-Bildung und -Rekrutierung ist wichtig für die Identifizierung potenzieller Ziele antiviraler Maßnahmen.
Publikation: Saenger L., Williams HM, Yu D., Vogel D. Kosinski J, Rosenthal M*, Uetrecht C*. An RNA to rule them all: Critical steps in Lassa virus ribonucleoparticle assembly and recruitment. BioRxiv DOI: 10.1101/2023.02.09.527830
Biochemische Charakterisierung des Lassa-Virus L-Proteins
Für unsere Untersuchungen produzieren wir das L-Protein des LASV in Insektenzellen mit Hilfe von Insektenviren. So können wir auf einer grundlegenden molekularen Ebene untersuchen, wie dieses multifunktionelle Multidomänenprotein das virale Genom vervielfältigt und die virale mRNA produziert. Wir haben verschiedene biochemische Tests entwickelt, die es uns ermöglichen, die vielfältigen Funktionen des L-Proteins zu untersuchen.
Das L-Protein synthetisiert RNA - dieser Prozess kann sowohl mit Hilfe von kurzen RNA-Fragmenten (Primern) gestartet werden als auch ohne Primer (de novo). Die RNA-Synthese benötigt Mangan-Ionen anstatt Magnesium-Ionen, wie es bei anderen Polymerasen üblich ist. Experimente mit unterschiedlichen kurzen Primern ergaben, dass es zwei Stellen für den Start der RNA-Synthese auf dem viralen Genom gibt und dass kurze RNA-Fragmente auf dem Matritzenstrang verschoben werden können. Diese Strategie wird als sogenannte „Prime-and-Realign“-Hypothese für die de novo Initiierung der RNA-Synthese vermutet. Außerdem konnten wir zeigen, dass das LASV Z-Protein die RNA-Produktion hemmen kann.
Publikation:
Vogel D, Rosenthal M, Gogrefe N, Reindl S, Günther S. Biochemical characterization of the Lassa virus L protein. J Biol Chem. 2019 Mar 29. pii: jbc.RA118.006973. doi: 10.1074/jbc.RA118.006973
Kouba T, Vogel D, Thorkelsson SR, Quemin ERJ, Williams HM, Milewski M, Busch C, Günther S, Grünewald K, Rosenthal M*, Cusack S*. Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat Commun. 2021 Dec 2;12(1):7018. doi: 10.1038/s41467-021-27305-5
Konformationsänderungen des Lassa-Virus L-Proteins während Promotor-Bindung und RNA-Synthese
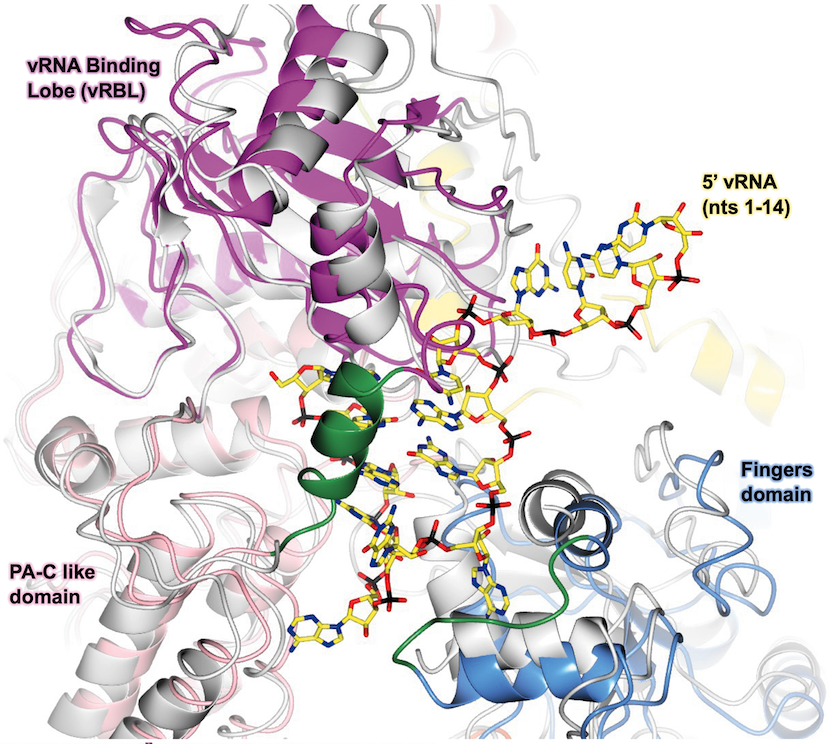
Wir konnten neun 3D-Strukturen des Lassa-Virus (LASV) L-Proteins mit Hilfe der Kryo-Elektronenmikroskopie bestimmen. Jede Struktur zeigt das LASV-L-Protein in einem anderen Bewegungszustand (Konformation), was die nötige Flexibilität dieses Moleküle für seine vielfältigen Funktionen unterstreicht. Diese hohe Flexibilität ist auch der Grund, warum die Strukturbestimmung so schwierig war. Da sich flexible Bereiche relativ schnell bewegen, sind sie sehr schwer abzubilden und erscheinen verschwommen, wenn sie überhaupt sichtbar sind. Unser Wissen aus der vorangegangenen biochemischen Charakterisierung des LASV L-Proteins ermöglichte es uns, das L-Protein in mehreren Funktionszuständen zu stabilisieren, die während der Genomvervielfältigung relevant sind. Wir konnten hochaufgelöste Strukturinformationen darüber erhalten, wie das LASV L-Protein durch Wechselwirkungen mit der viralen RNA reguliert wird. Wir haben verschiedene Bindetaschen für die konservierten 3'- und 5'-Enden der Promotor-RNAs festgestellt und gezeigt, wie die vollständige Bindung des RNA-Promotors eine bestimmte Konformation hervorruft, bei der das 3'-Ende in die aktive Tasche der RNA-abhängigen RNA-Polymerase (RdRP) zeigt. Während der frühen RNA-Elongationsphase und in Abwesenheit von viraler RNA wird die Endonukleaseaktivität des L-Proteins durch zwei verschiedene Peptide aus dem L-Protein gehemmt, während sie im Präinitiationsstadium nicht gehemmt wird. Im frühen Elongationsstadium ist ein RNA-Doppelstrang aus Matritzen-RNA und RNA-Produkt zusammen mit einem nicht hydrolysierbaren Nukleotid im aktiven Zentrum der Polymerase gebunden. Der gesamte C-terminale Bereich des L-Proteins, einschließlich der vermuteten Cap-Bindedomäne, ist dabei klar sichtbar. Diese Forschungsdaten verbessern unser mechanistisches Verständnis davon, wie dieses flexible und multifunktionale Protein aktiviert und reguliert wird.
Strukturelle und funktionelle Erkenntnisse über das L-Protein des Schweres-Fieber-mit-Thrombozytopenie-Syndrom-Virus
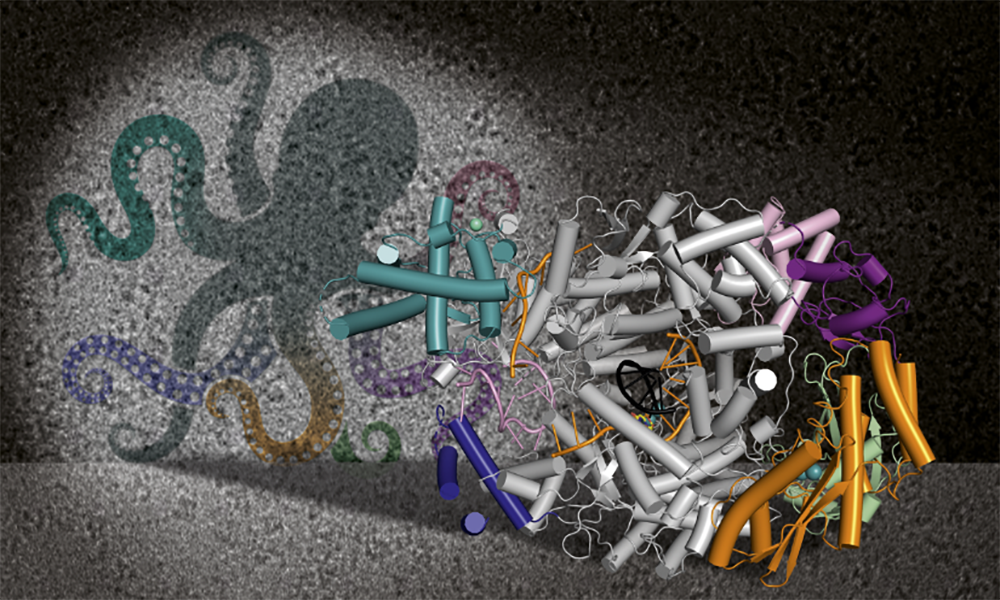
Im Jahr 2020 haben wir die erste 3D Struktur des L-Proteins des Schweres-Fieber-mit-Thrombozytopenie-Syndrom-Virus (engl.: Severe fever with thrombocytopenia syndrome virus, SFTSV) mit Hilfe der Kryo-Elektronenmikroskopie (Kryo-EM) bestimmt. Diese erste Struktur ermöglichte es uns, die grundlegende Architektur des SFTSV L-Proteins zu bestimmen, das mehrere funktionelle Domänen besitzt, darunter eine N-terminale Endonuklease, eine RNA-abhängige RNA-Polymerase (engl.: RNA-dependent RNA-polymerase, RdRp) und eine C-terminale Cap-RNA-Bindungsdomäne. Anschließend haben wir mittels Kryo-EM eine Reihe von hochauflösenden Strukturen des SFTSV-L-Proteins aufgeklärt, die verschiedene Phasen der Genomvervielfältigung darstellen, von der Prä-Initiation bis zur späten RNA-Elongation. So konnten wir zeigen, wie das SFTSV L-Protein die 5′-Virus-RNA in der Form eines Haken bindet (siehe Abbildung) und wie die distalen 5′- und 3′-RNA-Termini einen RNA-Duplex bilden, der das 3′-RNA-Ende im aktiven Zentrum der RdRp positioniert, bereit für die Initiation. Wir haben zudem Modelle des SFTSV L-Protein bestimmt, bei denen diese molekulare Maschine zuvor in der frühen und späten Elongationsphase gestoppt wurde. Dadurch konnten wir die Kanäle zu identifizieren, durch welch die neu synthetisierte RNA sowie die Matrizen-RNA das L-Protein verlassen.
Publikationen:
Vogel, D., Thorkelsson, S.R., Quemin, E.R.J., Meier, K., Kouba, T., Gogrefe, N., Busch, C., Reindl, S., Günther, S., Cusack, S., Grünewald, K., and Rosenthal, M., 2020. Structural and functional characterisation of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Research, doi: 10.1093/nar/gkaa253
Williams, H.M., Thorkelsson, S.R., Vogel, D., Milewski, M., Busch, C., Cusack, S., Grünewald, K., Quemin, E.R.J., and Rosenthal, M., 2023. Structural insights into genome replication by the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Research, doi: 10.1093/nar/gkac1249
Strukturelle und funktionelle Charakterisierung des Sin-Nombre-Virus-L-Proteins
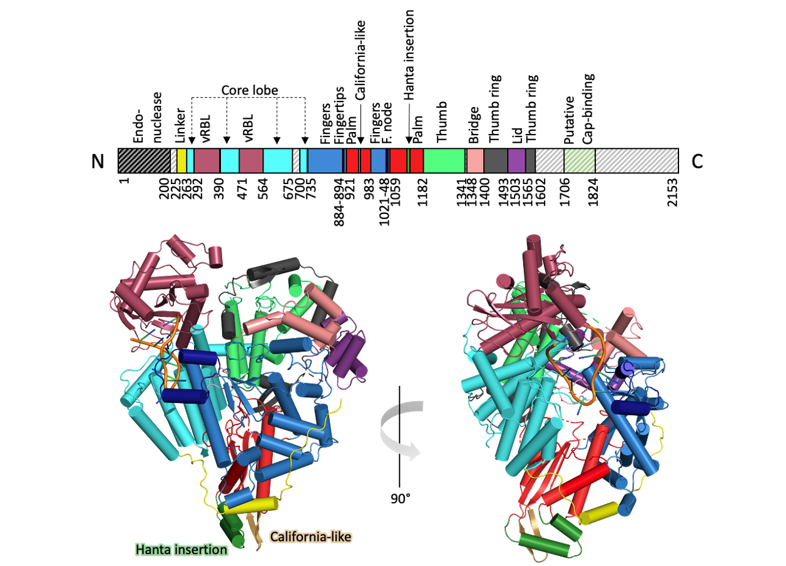
Hantaviren sind zoonotische Viren, die weltweit verbreitet sind und beim Menschen schwere Krankheiten verursachen können. Ein bedeutendes Beispiel ist das Sin-Nombre-Virus (SNV), welches das in Nordamerika am weitesten verbreitete humanpathogene Hantavirus ist. Bis heute gibt es keine wirksamen Impfstoffe oder spezifische, von der FDA zugelassene Therapeutika für die Behandlung von Hantavirus-Infektionen. Die gezielte Suche nach Hemmstoffen für die virale Genomvervielfältigung und Transkription erfordert eine tiefgreifende Kenntnis sowohl der Struktur als auch der Funktion des multifunktionalen L-Proteins. Wir haben Protokolle für die Expression und Reinigung des SNV L-Proteins sowie verschiedene biochemische Tests zur Untersuchung seiner Schlüsselfunktionen etabliert: (i) RNA-Abbau, katalysiert durch die Endonuklease, (ii) Bindung an die virale RNA und (iii) RNA-Synthese, katalysiert durch die RdRp. Darüber hinaus haben wir mittels Kryo-EM ein Strukturmodell des L-Proteins, einschließlich der RdRp, im Komplex mit dem viralen 5′-RNA-Ende erstellt.
Publikationen:
Meier K, Thorkelsson SR, Durieux Trouilleton Q, Vogel D, Yu D, Kosinski J, Cusack S, Malet H, Grünewald K, Quemin ERJ, Rosenthal M. Structural and functional characterization of the Sin Nombre virus L protein. PLoS Pathog. 2023 Aug 7;19(8):e1011533. doi: 10.1371/journal.ppat.1011533.
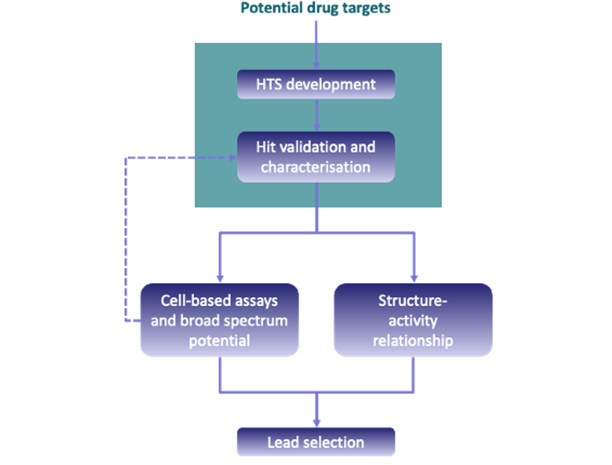
Umsetzung von Erkenntnissen aus der Grundlagenforschung in antivirale Strategien
Mit Hilfe von Kryo-EM haben wir mehrere L-Protein Strukturen im Komplex mit viraler RNA bestimmt. Innerhalb des L-Protein:RNA-Komplexes haben wir die verschiedenen Funktionen mithilfe biochemischer Assays und zellbasierter Studien charakterisiert, um regulatorische Elemente zu identifizieren. Wir testen nun die Eignung der verschiedenen L-Protein Funktionen und Regulationsmechanismen, wie beispielsweise die RdRp, RNA-Bindungsstellen und die Cap-Bindungsstelle, als potenzielle Breitband-Medikamentenziele. Zu diesem Zweck entwickeln wir unterschiedliche, auch hochdurchsatz-fähigeTestverfahren für das Screening kleiner Molekülbibliotheken. Wir arbeiten dafür mit dem Fraunhofer-Institut für Translationale Medizin und Pharmakologie (ITMP), ScreeningPort Hamburg, zusammen und haben Zugang zu Hochdurchsatztechnologien sowie großen Bibliotheken aus niedermolekularen Substanzen. Potenzielle Hit-Substanzen werden validiert und anschließend in zellbasierten Assays weiter charakterisiert. Parallel dazu werden die Treffer auf ihre Struktur-Wirkungs-Beziehung hin analysiert. Zu den verwendeten Methoden gehören Fluoreszenzpolarisation, homogener zeitaufgelöster FRET, Oberflächenplasmonenresonanz, Mikro-Thermophorese und Replikon-Assays. Mit diesem Projekt wenden wir Wissen aus der akademischen Grundlagenforschung des BNITM an um zusammen mit unseren Partnern am Fraunhofer ITMP neue antivirale Strategien zu entwickeln.