Genome and phylogeny
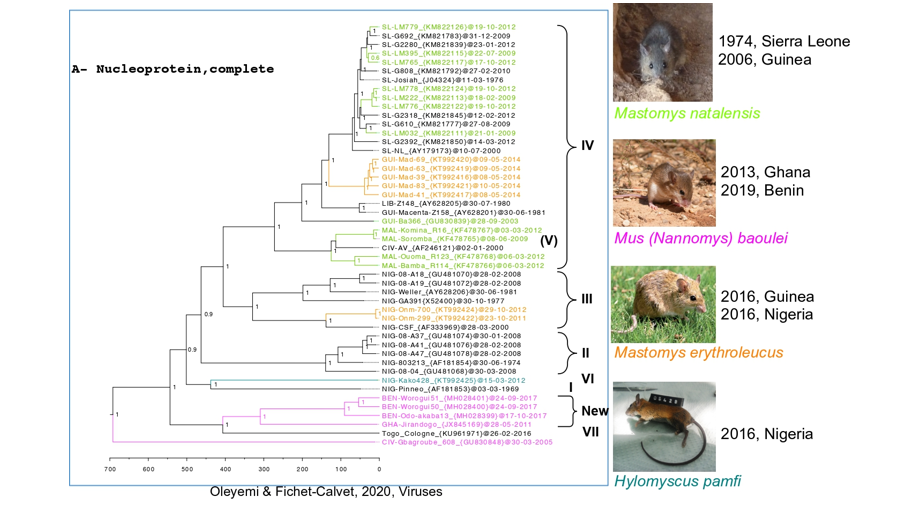
The genome of LASV is composed by 2 ambisense segments: the smaller segment (S) codes for the glycoprotein precursor and the nucleoprotein, and the larger segment (L) codes for the polymerase and the matrix protein. The coding regions are separated by an intergenic region with a strong secondary structure, and are flanked by non-coding regions. In 2000, Bowen et al. proposed a classification of the different strains with a phylogeny based on the GP, NP and polymerase. They suggested the existence of four lineages; 3 in Nigeria and 1 in the Sierra Leone-Liberia-Guinea.
Discovery of new hosts
In 2014, we discovered two new hosts: Mastomys erythroleucus in Guinea and in Nigeria, and Hylomyscus pamfi in Nigeria. In 2013 and 2019, we described the pygmy mouse, Mus (Nannomys) baoulei, as a possible carrier of a new LASV lineage in Ghana and in Benin.
Discovery of new lineages
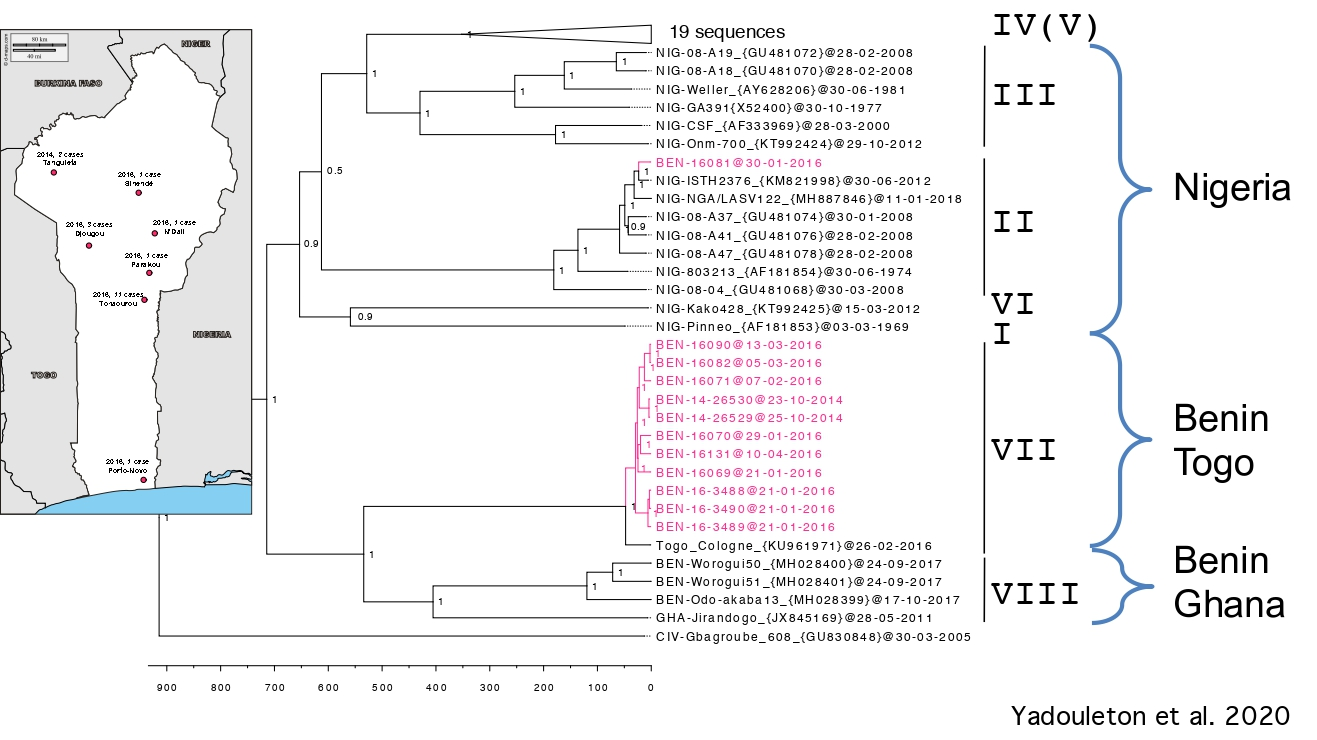
Investigations developed at ISTH in Nigeria revealed that some individuals had been infected with the Kako strain discovered in H. pamfi. The Kako strain therefore constitutes a new lineage because it is very distant from the lineage I discovered in 1969. Moreover, the recent Lassa epidemics in Benin and Togo (2014-2018) have shown that the patients were infected by a new strain, which forms a distinct 7th lineage.
LASV evolution at a local scale
In Upper Guinea, a longitudinal survey has been performed during 10 years. Viruses circulating in a specific locality are diverse and polyphyletic with respect to viruses from neighbouring villages. However, there are monophyletic clusters formed by viruses from a village at specific points in time, indicating that the temporal and spatial pattern of LASV evolution in the natural reservoir is characterized by a combination of stationary circulation within a village and virus movement between villages.
LASV emergence per host: human versus rodent
Here, we seek to phylogenetically infer ancestry and descent between LASV sequences detected in rodents and humans in selected localities within West Africa in order to provide increased insight into virus transmission at the rodent-human boundary.
Our aim is to compile a dataset including LASV sequences collected from both humans and Mastomys rodents within distinctive hotspots of Lassa fever.
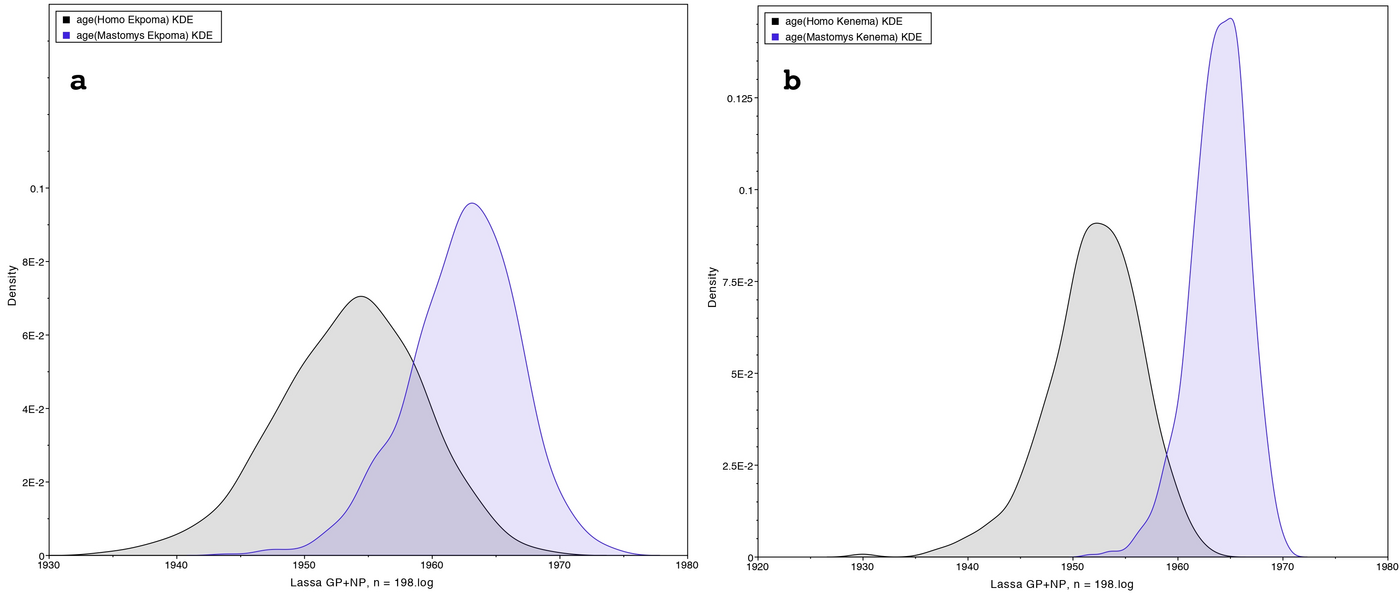
In a first study we phylogenetically compared LASV sequences obtained in two localities: Ekpoma in Nigeria and Kenema in Sierra Leone. We performed a time-calibrated phylogeny, using a Bayesian analysis on 198 taxa including 102 sequences from rodents and 96 from humans. Contrary to expectation, our results show that LASV strains detected in humans within these localities, even those sampled recently, are consistently ancient to those circulating in rodents in the same area.
Therefore, our results may suggest the occurrence of human-to-rodent transmission (reverse zoonosis).
Related key publications
Circulation of Lassa virus across the endemic Edo-Ondo axis, Nigeria, with cross-species transmission between multimammate mice
- Adesina AS, Oyeyiola A, Obadare A, Igbokwe J, Abejegah C, Akhilomen P, Bangura U, Asogun D, Tobin E, Ayodeji O, Osoniyi O, Davis C, Thomson EC, Pahlmann M, Günther S, Fichet-Calvet E, Olayemi A
- Emerg Microbes Infect. 2023 Dec;12(1):2219350. doi: 10.1080/22221751.2023.2219350
Lassa fever in Benin: description of the 2014 and 2016 epidemics and genetic characterization of a new Lassa virus
- Yadouleton A, Picard C, Rieger T, Loko F, Cadar D, Kouthon EC, Job EO, Bankolé H, Oestereich L, Gbaguidi F, Pahlman M, Becker-Ziaja B, Journeaux A, Pannetier D, Mély S, Mundweiler S, Thomas D, Kohossi L, Saizonou R, Kakaï CG, Da Silva M, Kossoubedie S, Kakonku AL, M'Pelé P, Gunther S, Baize S, Fichet-Calvet E
- Emerg Microbes Infect. 2020 Dec;9(1):1761-1770. doi: 10.1080/22221751.2020.1796528
Systematics, Ecology, and Host Switching: Attributes Affecting Emergence of the Lassa Virus in Rodents across Western Africa
- Olayemi A, Fichet-Calvet E
- Viruses. 2020 Mar 14;12(3):312. doi: 10.3390/v12030312
Determining Ancestry between Rodent- and Human-Derived Virus Sequences in Endemic Foci: Towards a More Integral Molecular Epidemiology of Lassa Fever within West Africa
- Olayemi A, Adesina AS, Strecker T, Magassouba N, Fichet-Calvet E
- Biology (Basel). 2020 Feb 7;9(2):26. doi: 10.3390/biology9020026.
Lassa Virus in Pygmy Mice, Benin, 2016-2017
- Yadouleton A, Agolinou A, Kourouma F, Saizonou R, Pahlmann M, Bedié SK, Bankolé H, Becker-Ziaja B, Gbaguidi F, Thielebein A, Magassouba N, Duraffour S, Baptiste JP, Günther S, Fichet-Calvet E
- Emerg Infect Dis. 2019 Oct;25(10):1977-1979. doi: 10.3201/eid2510.180523
New Hosts of The Lassa Virus
- Olayemi A, Cadar D, Magassouba N, Obadare A, Kourouma F, Oyeyiola A, Fasogbon S, Igbokwe J, Rieger T, Bockholt S, Jérôme H, Schmidt-Chanasit J, Garigliany M, Lorenzen S, Igbahenah F, Fichet JN, Ortsega D, Omilabu S, Günther S, Fichet-Calvet E
- Sci Rep. 2016 May 3;6:25280. doi: 10.1038/srep25280
Arenavirus Diversity and Phylogeography of Mastomys natalensis Rodents, Nigeria
- Olayemi A, Obadare A, Oyeyiola A, Igbokwe J, Fasogbon A, Igbahenah F, Ortsega D, Asogun D, Umeh P, Vakkai I, Abejegah C, Pahlman M, Becker-Ziaja B, Günther S, Fichet-Calvet E
- Emerg Infect Dis. 2016 Apr;22(4):694-7. doi: 10.3201/eid2204.150155.
Spatial and temporal evolution of Lassa virus in the natural host population in Upper Guinea
- Fichet-Calvet E, Ölschläger S, Strecker T, Koivogui L, Becker-Ziaja B, Camara AB, Soropogui B, Magassouba N, Günther S
- Sci Rep. 2016 Feb 25;6:21977. doi:10.1038/srep21977
Two novel arenaviruses detected in pygmy mice, Ghana
- Kronmann KC, Nimo-Paintsil S, Guirguis F, Kronmann LC, Bonney K, Obiri-Danso K, Ampofo W, Fichet-Calvet E
- Emerg Infect Dis. 2013 Nov;19(11):1832-5. doi: 10.3201/eid1911.121491









