DESY X-ray lightsource identifies promising candidates for COVID drugs
Joint major project in the fight against SARS-CoV-2
Hamburg. At the high-brilliance X-ray light source PETRA III of the German Electron Synchrotron (DESY), a research team in which the Bernhard Nocht Institute for Tropical Medicine (BNITM) is also active has identified several candidates for active agents against the coronavirus SARS-CoV-2. The promising drug candidates bind to an important protein of the virus and could thus be the basis for a drug against COVID-19.
In a so-called X-ray screening, the researchers under the leadership of DESY quickly tested almost 6,000 active substances that already exist for the treatment of other diseases. After measuring around 7,000 samples, the team was able to identify a total of 37 substances that bind to the main protease (Mpro) of the SARS-CoV-2 virus, as the scientists report online today in the renowned journal "Science". Seven of these substances in turn inhibit the activity of the main protease and thus slow down the multiplication of the virus, as the BNITM found out. Two of them are so promising that they are currently being further investigated in preclinical studies. What is probably the largest drug screening of its kind also brought to light a new binding site on the main protease of the virus to which drugs can couple.
![Direct view of the sample in protein X-ray crystallography [Translate to English:] Das Bild zeigt eine blau leuchtende Messapparatur.](/fileadmin/media/Aktuelles/2021/DESY_Wirkstoffscreening_1071_540x330.jpg)
In contrast to vaccines, which help healthy people to defend themselves against the virus, drug research is looking for drugs that slow down or stop the reproduction of the virus in the body of sick people. Viruses cannot reproduce on their own. Instead, they introduce their own genetic material into the cells of their host and cause them to produce new viruses. Proteins such as the main protease of the virus play an important role in this process. It cuts protein chains, which were produced by the host cell according to the blueprint of the virus genetic material, into smaller parts that are necessary for the reproduction of the virus. If the main protease can be blocked, the cycle can be interrupted under certain circumstances; the virus can no longer reproduce and the infection comes to a standstill.
High-tech in structural biology fro thousends if candidates to the best two
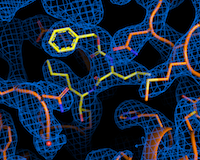
The P11 beamline of DESY's PETRA III research light source specialises in structural biology studies. Here, the three-dimensional spatial structure of proteins can be displayed with atomic precision. The research team led by DESY physicist Alke Meents used this to examine several thousand known active substances from a library of the Fraunhofer Institute for Translational Medicine and Pharmacology and another of the Italian company Dompé Farmaceutici SpA to see whether and how they "dock" to the main protease - the first important step in blocking it. Like a key in a lock, the drug molecule fits into a binding centre of the protease. The advantage of the drug library is that it contains active substances that have already been approved for the treatment of humans or those that are currently in various testing phases. Suitable candidates to combat SARS-CoV-2 could therefore be used in clinical trials much more quickly, saving months or years of drug development.
The special technical equipment at the PETRA III station P11 includes a fully automated sample change with a robotic arm, so that each of the more than 7,000 measurements took only about three minutes. With the help of automated data analysis, the team was able to quickly separate the wheat from the chaff. "Using a high-throughput method, we were able to find a total of 37 active substances that bind with the main protease," says Meents, who initiated the experiments.
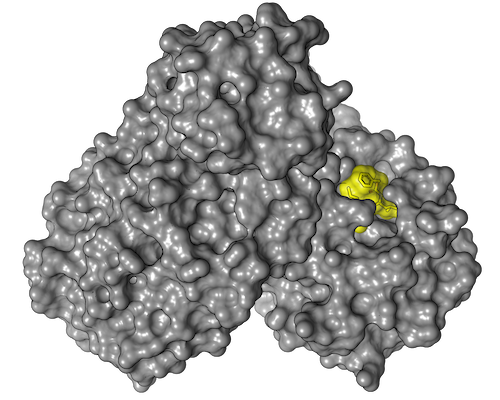
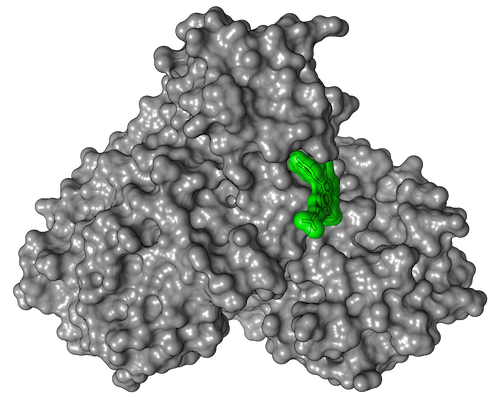
In a next step, the research group led by Yaiza Fernández-García from the Department of Virology at BNITM investigated whether these agents inhibit or even prevent virus replication in cell cultures and how compatible they are for the host cells. This reduced the number of suitable active substances to seven, two of which stood out in particular. The active substances calpeptin and pelitinib clearly showed the highest antiviral activity with good cell compatibility. Therefore, cooperation partners have already started preclinical investigations with these two active substances.
New binding site identified
In their drug screening using protein crystallography, the researchers did not examine fragments of potential drugs as usual, but complete drug molecules. In the process, the team of more than 100 scientists also discovered something completely unexpected: they found a binding site on the main protease that had been completely unknown until then. "It was not only a positive surprise that we were able to discover a new drug binding site on the main protease - a result that can really only be achieved at a synchrotron light source like PETRA III - but that even one of the two hot drug candidates binds precisely to this site," says Christian Betzel from the University of Hamburg, co-initiator of the study.
A particular strength of the X-ray screening method compared to other screening methods is that researchers obtain the three-dimensional structure of the protein-drug complexes as a result and can thus determine the binding of the drugs to the protein at the atomic level. Even if the two most promising candidates do not make it into clinical trials, the 37 substances that bind to the main protease form a central database for drug developments based on them.
Background information
Original publikation: X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease; Sebastian Günther, Patrick Y. A. Reinke, et al.; „Science“, 2021; DOI: 10.1126/science.abf7945
Official press release : DESY News 02.04.2021
The cooperation partners: In addition to DESY and BNITM scientists, researchers from the Universities of Hamburg and Lübeck, the Fraunhofer Institute for Translational Medicine and Pharmacology, the Heinrich Pette Institute, the European XFEL, the European Molecular Biology Laboratory EMBL, the Max Planck Society, the Helmholtz Centre Berlin and other institutions are involved in the work. In addition to the experiments at the P11 measuring station, measurements were also carried out at the EMBL measuring stations P13 and P14 at PETRA III.
Contact person
PhD Yaiza Fernández García
Research Group Leader
Phone : +49 40 285380-942
Email : yaiza.fernández-garcía@bnitm.de
Prof. Dr Stephan Günther
Head of Department of Virology
Phone : +49 40 285380-547
Fax : +49 40 285380-459
Email : guenther@bnitm.de
Dr Eleonora Schoenherr
Public Relations
Phone : +49 40 285380-269
Email : presse@bnitm.de
Julia Rauner
Public Relations
Phone : +49 40 285380-264
Email : presse@bnitm.de






